Collaboratories for Genetics Learning
Genetics data stores have grown explosively in size and complexity over the past two decades. The amount of public DNA sequence data available has doubled every 14 months for 20 years.1 A technique announced in January 2009 promises to make DNA sequencing 30,000 times faster, sequencing an entire human genome in less than 30 minutes for under $1,000.2 This technique will be available for doctors and patients within two years.3 These rapid developments in genetics will radically affect individual health decisions, national political debates, and human ethical concerns.
Open any textbook, however, and you’ll read a different story entirely. In current classroom materials, genetics looks far too much like a dusty science of the nineteenth century. Students first encounter this modern science by reading about a monk studying wrinkled peas a century and a half ago. Historical perspectives are important in understanding modern biology, but current curricula rarely do justice to biology’s recent transformations. Entire new fields within genetics and DNA science have appeared in the period since existing national science curriculum standards were published, and Mendel’s rules from the 1800s have been exposed as very narrow exceptions to the true, complex process of inheritance.
Biology isn’t the only thing in flux. Technology, connectivity, and the need to tackle complex problems have made science more collaborative than ever. Scientists work in global “collaboratories”—collaborative laboratories—to combine expertise across disciplines. For many, the computer has replaced the lab bench as the main experimental tool for the new discipline of bioinformatics, the application of statistics and computer science to the study of molecular biology. Modern techniques and analyses rely on databases that are populated with the findings of thousands of researchers across the globe.
Current classroom approaches fail to expose students to current content and techniques, but even more importantly, they do not capture the dynamic, collaborative nature of science. Tomorrow’s scientists will enter future professions armed with an obsolete education.
This problem calls for more than a simple fix. Every year, textbook publishers tuck a few new pages into existing thousand-page tomes or create sidebars describing new scientific discoveries, but this treatment does little more than pay lip service to the emerging sciences and fields of study. The knowledge is also different enough that traditional teaching approaches simply do not work. In order to help students understand the complex world of genetics and genomics and appreciate their intricate details, curricula must reach much deeper and engage students in a new manner. The Geniverse project, funded by the National Science Foundation, is building such a curriculum.
Enter the world of dragons
Geniverse extends an existing partnership among the Concord Consortium, the Maine Mathematics and Science Alliance, and the Jackson Laboratory, and adds education researchers from BSCS. Building on an exploratory project that brought cutting-edge genomics experimental techniques to high school students, Geniverse is creating a collaborative web-based environment in which students study core genetics and heredity concepts, collect evidence and study a problem, and work together as scientists to solve relevant scientific puzzles and challenges.
Geniverse shares a pedigree with past Concord Consortium projects reaching back to the pioneering GenScope software and is built upon the same compelling premise—students explore relevant aspects of heredity and genetics by breeding and studying virtual dragons.
These dragons have made it possible to link genotype and phenotype visually, bringing vital biological connections to life for students. Using this software, students can quickly grasp the connection between genes and traits and easily understand the way Mendelian inheritance works. (For example, the genotypes Hh and HH both express the phenotype graphically as a dragon with horns while a dragon with the recessive hh does not display horns.) The Geniverse project is extending these capabilities; dragons’ traits are now linked to the molecular nature of genes. Further, students are able to collaborate to ask and answer important questions.
The tools allow students to move quickly and easily among three critical levels of biological investigation and understanding: the nano-level (nucleotides), micro-level (chromosomes), and macro-level (traits). As they do so, students make important conceptual connections among these integral stages. Narrative scenarios woven through the curriculum engage students and ensure that the connections between these levels are relevant and compelling. For example, in one scenario, while breeding dragons a student may uncover a disease and learn that it is spreading through the population unchecked.
The student encountering this disease is able to analyze the affected dragon’s DNA and compare it with known sequences in an online database, identifying a specific mutation as the cause of the disease. Instructional sequences such as this require students to apply analytical skills to this multi-level model and perform virtual laboratory procedures in attempts to pinpoint the difficulties causing the disease and to treat it.
Moving to a more complex understanding
A student who completes a Geniverse curriculum module may have correctly identified a recessive mutation as a cause of the disease in the dragon population. Upon concluding this, she can share her result with the rest of the class as a “publication.” As part of her publication, she also shares the line of dragons she has isolated as evidence supporting her discovery. Other students in the class test her research by breeding their own dragons with her isolated strain and verifying her result.
Soon after this, however, the same student might learn of a publication by another student that clearly contradicts her findings. This student has bred the isolated strain with a disease-bearing strain of his own. When the two diseased strains are bred together, the offspring are mysteriously disease-free. What can be causing this strange result?
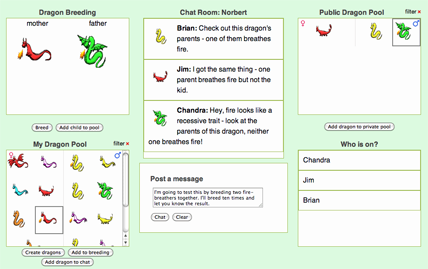
To further understand the problem, students must work together, share findings and evidence, cite other results, and share database searches, dragons, and DNA snippets. Through their collaboration, it gradually becomes clear that two separate mutations in entirely different genes can cause the same disease. In fact, the students discover, the two genes both act in a single pathway—a sequence of enzyme-powered, biochemical reactions that is essential for health; if any gene in the pathway is faulty, the pathway fails, resulting in the same disease.
While solving a mystery, students uncover for themselves the underlying nature of gene expression: by virtue of the molecules they encode, genes act in concert, and not simply as individual entities. Students move beyond “dominant” and “recessive” and enter a more complex, multilayered understanding of genetics.
In other modules, students work both as individual learners and collaborating scientists to solve additional challenges, revealing and addressing various quandaries in the dragon population while building and reinforcing their understanding of core genetics concepts. By layering such puzzles and compelling narratives within standard curriculum, the Geniverse project exposes students to the techniques of modern genetics and helps students develop an understanding of biology as an active, experiment-driven science. With a solid foundation of core genetics concepts, students will be better prepared for a future in which these concepts are increasingly important.
Frieda Reichsman (freichsman@concord.org) directs the Geniverse project.
Chad Dorsey (cdorsey@concord.org) is President of the Concord Consortium.