Students Learn Genetics with Geniventure
Our mission is to ignite large-scale improvements in teaching and learning through technology, so we’re especially interested in how our free educational technologies fare in the classroom after the funded projects that produced them have ended. Recently, we have been studying students’ learning of genetics through their interactions with Geniventure, an immersive digital game designed for middle and high school students that features a narrative about dragons.
In Geniventure, a war has broken out between kingdoms, endangering the dragon population. Each player breeds drakes—the model species for dragons—to help win the war and prevent dragon extinction. A diverse cast of characters in a scientific guild presents 65 challenges that require students to learn about drake genetics. As they progress through the game, the software logs each student’s actions. Some actions are productive, moving the student closer to the goal, some are counterproductive, and some are simply redundant. Students’ ability to discriminate between these possibilities, informed by their understanding of the underlying genetics, can be estimated by examining their actions. We applied this strategy to the analysis of a subset of the challenges.
The target-match challenges
There are 22 target-match challenges, each with a common goal: to change the genes on a drake to make it look like a target drake, randomly generated at the start of the challenge. The challenges also set a subsidiary goal—to achieve a match with as few moves as possible, where a move is defined as either a change to a gene or a sex change. The minimum number of required moves is calculated by the game, based on the phenotype (the set of observable traits) of the target drake and the initial genotype (the genetic makeup) of the manipulable drake (Figure 1).
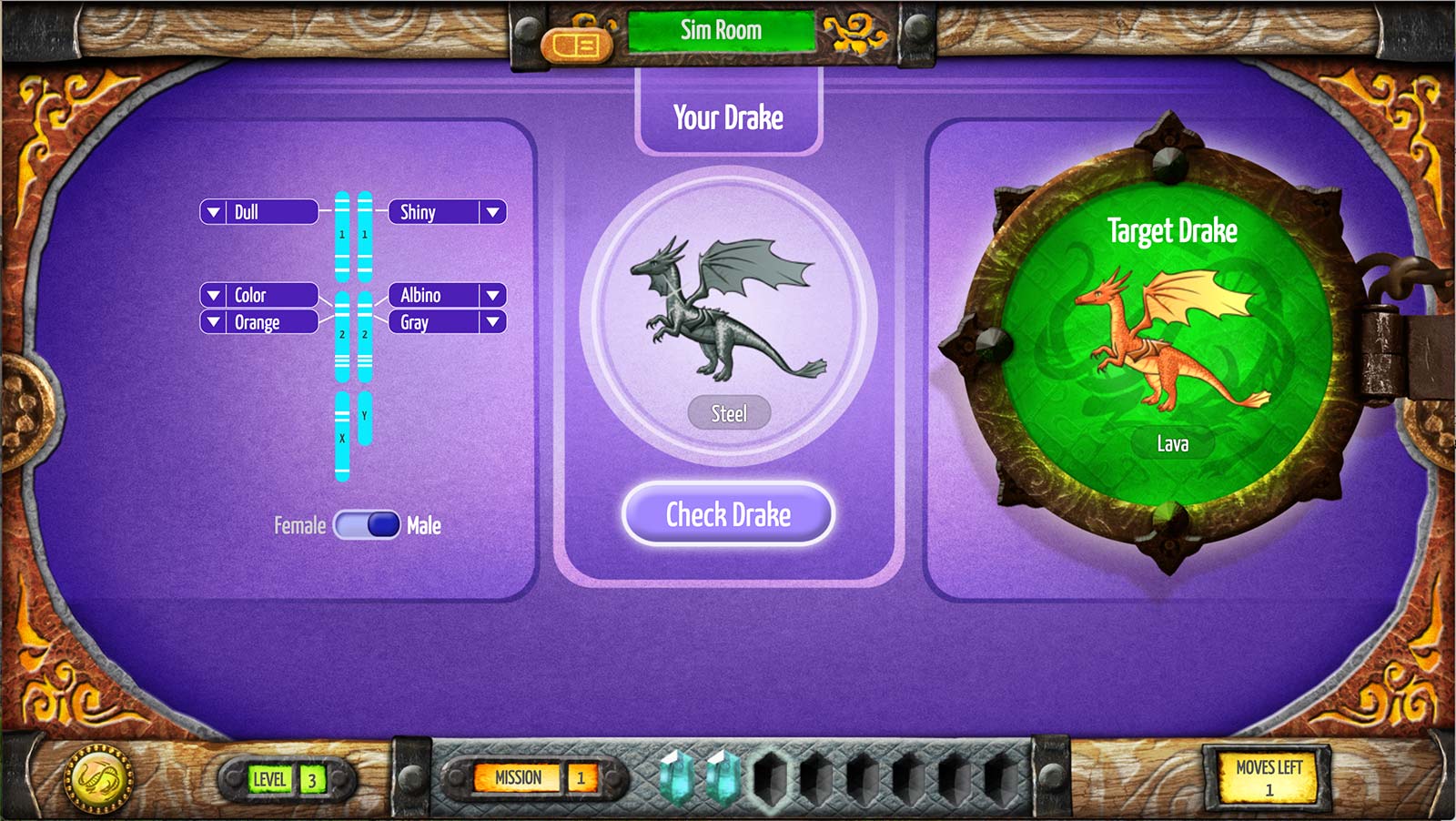
Students who reach the target state making no more than two moves over the minimum number are rewarded with a crystal, the color of which denotes how successful they were. Achieving a match with the minimum number of moves results in a blue crystal—the most prized of all. One excess move results in a yellow crystal; two excess moves and the crystal is red. Matching the target drake with more than two moves over the minimum is counted as a success, but students are not awarded a crystal. If the drakes do not match, the student continues the challenge with the same target drake; if they do match but the number of moves exceeds the minimum, the student is offered the chance to try to achieve a better crystal by repeating the challenge with a different target. Thus a single challenge can give rise to multiple attempts.
The target-match challenges occur in four clusters throughout the game, separated by challenges of other types. Each cluster starts with the manipulable drake continuously visible to the student, sometimes changing its appearance after a move in accordance with an underlying model of drake genetics. (For instance, if the student changes ww to Ww, the drake transforms from wingless to winged.) In this condition, students can make as many moves as they like, observing the effect each time, and only submitting their drake when it looks exactly like the target drake. The fact that they can immediately observe the effects of their moves encourages students to experiment in order to work out the mapping between genotype and phenotype.
After two or more challenges with a visible drake, the challenge changes to the “hidden” condition: the manipulable drake is represented by an unhatched egg and becomes visible only when the egg is submitted (Figure 2). In the hidden condition students must match the manipulable drake to the target just by observing its genes and applying their knowledge of the genotype-to-phenotype mapping.
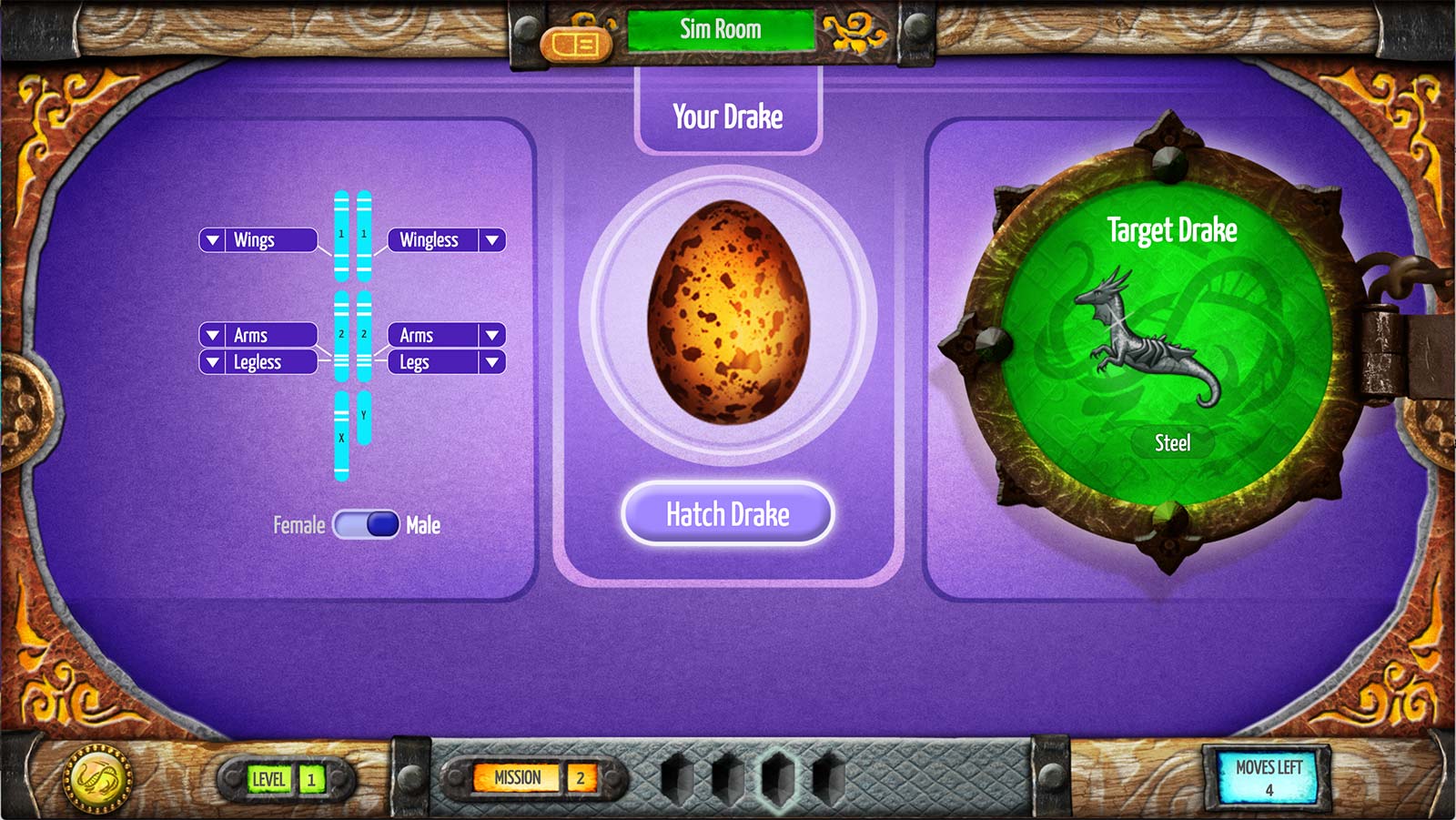
The visible-mode challenges are intended to prompt experimentation and inquiry, while the hidden challenges act as embedded assessments of content knowledge. The shift from visible to hidden conditions repeats in the four clusters of target-match challenges, each involving a different set of traits and a more complex mapping (for example, color is determined by more than one gene).
The target-match challenges are useful for inferring a student’s mental state because they are not limited to correct or incorrect matches but involve the sequence of actions that resulted in that outcome, actions that reflect a student’s understanding of genetics. To quantify students’ performance we developed and automated a scoring rubric that takes into account both the number of times a student attempts a challenge and the best crystal color awarded on that challenge.
Two research cohorts
In order to assess the effectiveness of our Geniventure game to teach genetics, we compared two cohorts of students taught by teachers who had different preparation. The first group of teachers took part in our research project and participated in face-to-face professional development while the second group of teachers registered independently and did not have additional training.
During the spring of 2019, the six teachers who participated in our National Science Foundation-funded GeniGUIDE research project implemented Geniventure in their classrooms. All had participated in a three-day workshop and were familiar with the game and its underlying science content and pedagogy. All administered identical multiple-choice tests to their students before and after they played the game. These students constitute our research cohort.
Later, between August 2019 and April 2020, 510 teachers outside our research project registered to use Geniventure. Of these, 22 administered both the pre- and post-test. Their students constitute our extended cohort.
After filtering out students who did not complete all the target-match challenges, as well as those who failed to answer at least 95% of the items on the pre- and post-tests, there were 338 students in the research cohort and 433 in the extended cohort. We matched the students’ scores on the post-test to their pre-test scores and their target-match challenge scores in both the visible and hidden conditions.
Findings
- Based on a two-tailed t-test, the means of the students’ post-test scores were significantly higher (p < .001) than the means of their pre-test scores. This was true for both cohorts. The effect size (Cohen’s d) was 0.83 for the research cohort and 0.57 for the extended cohort.
- Using pre-test score, visible-challenge score, and hidden-challenge score as the independent variables, the post-test score was significantly correlated (p < .001) with pre-test score and with the challenge score in the hidden condition but not in the visible condition. This was true for both cohorts.
- Adding cohort as an independent variable to the regression analysis did not significantly improve the fit. Thus, on these measures the two cohorts are statistically indistinguishable.
Discussion
Students in both cohorts improved their knowledge of genetics as measured by a multiple-choice assessment, evidence that the research project is having a broad impact. Their post-test scores correlate only with performance on the hidden target-match challenges, consistent with educational theory in that the visible challenges probe for inquiry skills rather than content knowledge. The significant correlation between in-game performance and summative test scores justifies using one as a proxy for the other. This will enable us to analyze log data generated by students who use Geniventure but do not take the pre- and post-tests, including those who use the software remotely. Next, we will investigate the extent to which the Geniventure materials may be successful even in the absence of face-to-face contact with a teacher.
Paul Horwitz (phorwitz@concord.org) is a senior scientist.
Trudi Lord (tlord@concord.org)is a senior project manager.
Frieda Reichsman (freichsman@concord.org)is a senior research scientist.
This material is based upon work supported by the National Science Foundation under grant DRL-1503311. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.