A Drake’s Tale: Genetics Software Gets a Lift from Gaming
Many of us learned about dominant and recessive genes in a humdrum high school biology class. Some of us may still recognize the terms and symbols twenty or thirty years later—are your eyes bb or Bb? But, as it turns out, a very small number of traits in humans and other animals, plants, amoeba … you name it … involve the dominance mechanism of a single gene with just two alleles. (An allele is a variation of a gene, like the B or b in the above example.) The more biologists discover about the mechanisms of inheritance, the fewer traits we can point to that involve only one gene or can be illustrated using a simple Punnett square. In fact, biologists are compiling information about our genes at an astounding rate. As the process of sequencing DNA improves, the science of biology is dramatically changing.
Modern biology showcases both the relatedness and complexity of genetic interactions across virtually all organisms. For example, you may be surprised to learn that humans and fruit flies share a core set of genes comprising 60% of their genomes, and further, that 90% of mouse and human genes are similar to each other. As for complexity, there are at least eight genes that control eye color, and by the way, two individuals with blue eyes can have a child with brown eyes. Today’s students may be the first with access to enough information to break out of the Punnett square box and explore the many different ways in which genes act and interact. Students who use our Geniverse software and curriculum attain the keys to unlock the mysteries of the genetic code. As they discover polyallelic, multigenic, and sex-linked traits, they develop a broader and deeper knowledge of genetics.
Playing the Game
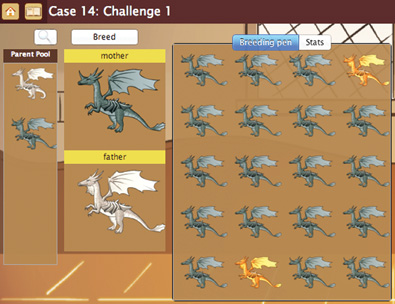
Geniverse immerses students in a game-like environment where they are challenged to sort out the genetics of a mythical organism, the drake (Figure 1). Drakes are essentially a smaller version of a dragon, and are a model species that can help solve genetic mysteries in dragons, in much the same way as the mouse is a model species for human genetic disease. In fact, the Concord Consortium began the exploration of the genetics of dragons with the GenScope project led by Paul Horwitz in 1992, followed by BioLogica in 1998, and then GENIQUEST in 2007. With Geniverse, we are adding elements of gaming to this engaging approach to learning genetics.

To become a Master breeder in the Drake Breeders Guild, students must learn the tricks of the trade to produce a variety of drakes, which they can do only by understanding the ways that drake genes interact and are inherited. Students explore the genetic landscape by doing experiments, looking at the data, drawing tentative conclusions, and then testing these conclusions with more experimentation. Thus, despite moving through a fantastical world, students are engaging in an authentic, experiment-driven approach to biology.
To progress through the Guild, each student solves a series of “cases,” moving from training level to Apprentice, Journeyman, and ultimately Master. By the time students reach the game’s finale, they have encountered genes that have more than two alleles, genes whose alleles are not fully dominant or recessive (known as incomplete dominance), genes that exhibit different patterns of inheritance in males and females (X-linked genes), and a trait that is influenced by three different genes.
Students learn that while drakes are mythical creatures, the drake genome comes from real-world animal models, including mouse, lizard (Figure 2), and stickleback fish. For example, tough exterior drake “armor” plates are encoded by a real gene involved in generating some typical properties of skin: in anole lizards, scales; in humans, hair and sweat glands. There are four alleles of the armor gene. Their pairwise combinations can yield from zero to five plates.

Armor illustrates that there are a variety of versions of a trait (phenotypes) that can be present in a population, even though any individual can only have two alleles (one from the mother and one from the father).
Both simple and complex traits are packed into the three pairs of drake chromosomes. Geniverse includes standard dominant/recessive traits (drake wings, forelimbs, and hindlimbs). Two trickier forms of dominance round out the collection of simpler traits: a rostral horn, which is a sex-linked trait, and horns on the top of the head, a recessive trait. The last one slays the common misconception that the presence of a trait is always the dominant condition.

As one of the final tests of their ability to uncover genetic mechanisms, students must fully explain the genetic control of the eight possible drake colors that result from the interaction of three genes. Students conduct breeding experiments both individually and in small groups (because not every student has access to drakes of every color), and publish their results in the Journal of Drake Genetics. Because this level of complexity goes beyond the usual learning goals for introductory biology, one might assume that student interest is lost or declines at this late stage of the game. But the opposite is true. One teacher described her students’ excitement: “My kids are pumped about color! The more they are learning, the more they can’t wait to figure out how this all works. It’s a great program!”
DNA and Genes
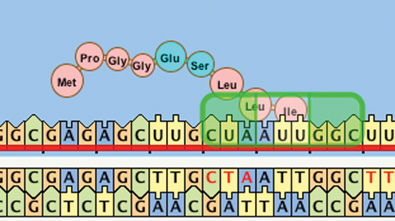
Current development of the Geniverse software involves illuminating the molecular path from genotype to phenotype, that is, from the DNA that makes up a gene to the trait it encodes. At strategic points in their fantastical journey, students use a simulation of meiosis — the process of formation of gametes (sperm and egg cells) — which enables them to follow the many possible reassortments of parental alleles into gametes. Some of the mechanisms of genetic inheritance and diversity are thus revealed. As students progress, they will delve deeper and deeper into the underlying genetic code, digging down to the DNA sequence level using a protein synthesis simulator (Figure 3) and a database of drake DNA sequences populated with real genes.
Students will make small changes in a gene sequence and learn how such seemingly minute changes can either drastically affect an organism’s physical appearance and metabolic functions or have no effect at all. When the Guild summons all drake breeders to help cure a genetic disease, students will use these tools to track down the cause of the disease.
Rather than memorizing terms and working through problems with selected data, students who play Geniverse learn to reason their way through a genetic challenge by crafting experiments that probe for information and answers, generating their own data sets and conclusions. The topics of inheritance, meiosis, and DNA-to- protein are frequently taught as separate units in traditional biology courses, making it difficult for students to connect their biology class to the real world. Geniverse helps teachers and students weave these topics into a meaningful tapestry, enabling students to better understand the big picture. And who doesn’t like playing with dragons?
Frieda Reichsman (friechsman@concord.org) directs the Geniverse project.
Trudi Lord (tlord@concord.org) is the Geniverse project manager.
This material is based upon work supported by the National Science Foundation under grant DUE-0733264. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of the National Science Foundation.