Molecular Modeling in Cyberspace
The 2013 Nobel Prize in Chemistry was awarded to Martin Karplus, Michael Levitt and Arieh Warshel “for the development of multiscale models for complex chemical systems.” According to the secretary of the Royal Swedish Academy of Sciences, “This year’s prize is about taking the chemical experiment to cyberspace.”
Our Molecular Workbench software was developed over the past decade by Senior Scientist Charles Xie, whose own postdoc co-advisor Georgios Archontis was advised by none other than the new Nobel Prize winner Martin Karplus! Georgios was an early contributor to CHARMM, a widely used computational chemistry package. Charles collaborated with both Georgios and Martin Karplus on a study that used CHARMM to calculate binding affinities among biomolecules involved in the metabolism of glycogen.
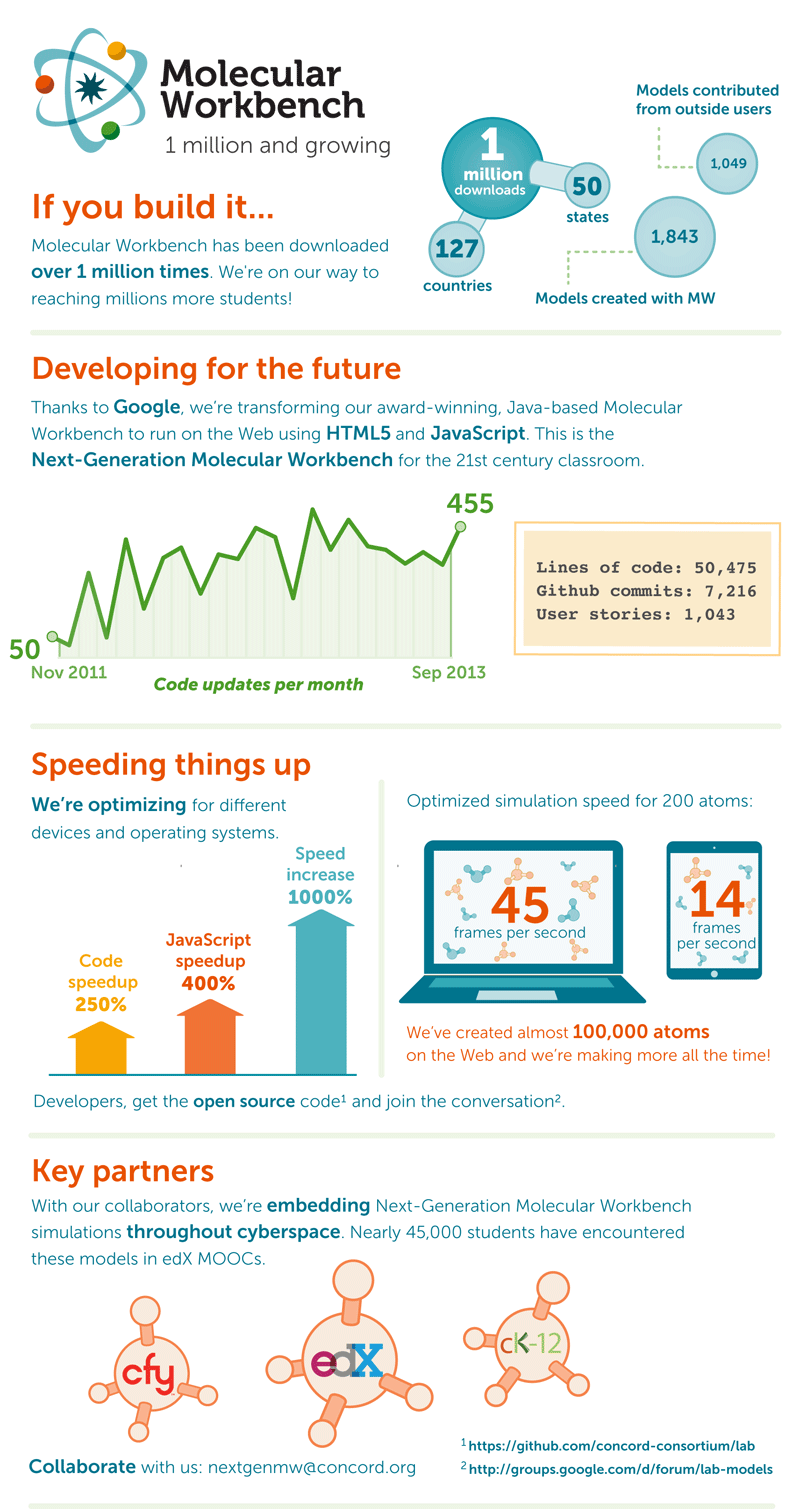
Molecular Workbench (MW) is built on the fundamental physical principles that govern molecular dynamics. This ensures the accuracy and depth of the visual simulations, and many of its computational kernels were inspired by CHARMM. We’re proud of this legacy and honored by the academic genealogy. Now, with funding from Google, we’re transforming the Java-based Classic MW software to run on the Web using HTML5 and JavaScript. With this, we’re taking the chemical experiment to cyberspace, this time for education.
A microscope for the molecular world
Understanding the world around us is an elusive prospect. From events as simple as melting ice and boiling water to those as subtle and complex as the interplay between light and matter, mysterious rules and invisible processes seem to rule every aspect of life. Understanding science demands a thorough familiarity with them. Or does it?
Formulas and algorithms are perfect for experts already familiar with them. To students, however, they are a foreign language. More often than not, they obscure the phenomena they attempt to describe instead of making them more accessible. But this doesn’t need to be the case. Models and simulations can make the invisible visible and explorable for students, turning a mysterious and obscure world into one that is tangible, conceptually available and ripe for student experimentation— all without a single equation.
Transforming Molecular Workbench for the web
Our Java-based Classic Molecular Workbench has been making this possible for students for nearly a decade. In 2011, Google awarded the Concord Consortium funding to bring MW software to the next stage. Over the past two years, our Next-Generation MW project has worked to bring the already popular Molecular Workbench to a wider audience via the power of the Web, with a goal of exposing real-world phenomena for student learning through exploration. We have created a series of tools that allow students to experience both microscopic and macroscopic phenomena in new ways. These are part of a new generation of learning— web-based and accessible on all manner of devices—that will permit millions of students around the globe to investigate scientific phenomena and help redefine teaching and learning for years to come.
Molecular Workbench brings a stunning range of phenomena to students’ laptops and tablets. Students can investigate fundamental molecular processes underlying everyday phenomena—from phase change to diffusion through membranes— and understand the variables that control them. Computational models unveil the interactions between light and matter, demonstrating how plants harvest light for photosynthesis or how different wavelengths of light interact with CO2 in the atmosphere. Models of DNA reveal how encoded information is translated into the proteins that govern processes in all living things and how small mutations in this code can cause large changes, or no changes at all. Demonstrations of basic chemical reactions show how a single spark and a series of random molecular motions can result in an explosive cascade of molecular changes.
Macroscopic phenomena governed by fundamental physical equations are equally accessible using these same modeling engines. Molecular Workbench makes physical phenomena such as the motion of a pendulum or the vibration of a mass on a spring available for intricate investigations, with linked graphs that help expose the subtleties behind these seemingly simple systems. And other simulation engines make even more phenomena accessible. Students can also explore the motion of the planets, governed by the very same equations but on a vastly grander scale. Simulations inspired by Charles Xie’s Energy2D software bring the world of fluid dynamics, energy and heat transfer alive, permitting students to see conduction, convection and radiation, and to perform experiments to understand engineering principles involved in concepts such as solar house design.
Simulations make these phenomena and many more available to students around the globe. Perhaps some day one of them will be inspired by Molecular Workbench to win a Nobel Prize in Chemistry—molecular modeling certainly has more awards in its future.
Chad Dorsey (cdorsey@concord.org) is President of the Concord Consortium.
Infographic design by Emily Marsh.
This material is based upon work supported by Google.org. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the author(s) and do not necessarily reflect the views of Google.org.