Secondary Students Break Into Genetics Research
Students crack the DNA code for mice and dragons.
Two student interns are bent intently over computer monitors. In a nearby building, tens of thousands of mice scurry in cages, each labeled with their specific genetic strain. Years of painstaking laboratory breeding have developed each strain’s unique characteristics. Some mice always become obese. Some are bald. Others have extra large ears. One kind even glows bright green under ultraviolet light. These famous mice are the reason students are here, studying genetics. Suddenly, one student gasps in surprise. Pointing to the image on her companion’s screen, she exclaims, “How did you get yours to breathe fire?”
A similar scene marked the early stages of the GENIQUEST project, an NSF Discovery Research K-12 (DR-K12) grant from the Maine Mathematics and Science Alliance in Augusta, Maine on which The Jackson Laboratory in Bar Harbor, Maine and the Concord Consortium are collaborators. Of course, the organisms on the computer screen weren’t a new strain of fire-breathing mice. Instead, they were the dragons that so many students have come to know through our BioLogica genetics software. The project brings cutting-edge genetics research techniques to high school students.
GENIQUEST stands for Genomics Inquiry through Quantitative Trait Loci Exploration with SAIL Technology. The project builds on Jackson Laboratory’s 75-year history of student summer interns and the lab’s recent success extending this outreach to magnet high schools such as the Maine School of Science and Mathematics and the North Carolina School of Science and Mathematics. Jackson Lab researchers mentor students in the latest genetics research techniques, including Quantitative Trait Loci (QTL) analysis, a statistical process used to pinpoint the location of unknown genes.
Breeding dragons
Students are transported to a mythical island where dragons live. They begin by breeding and studying the dragons to confirm the basic Mendelian genetics they have learned in the classroom. Mysteries crop up, however, when students discover a trait whose cause they can’t identify through the software. Some dragons develop plates on their upper neck, but the gene that controls this trait is not obvious.
To learn more, students “attend” a research conference on the island and read research papers from famous dragon scientists. They investigate the appearance of these mysterious plates by breeding individuals. Then they examine the offspring’s chromosomes, using their knowledge of the parent strains and information about the recombined DNA in the offspring to narrow down the possible location of the gene for neck plates. By simple manipulation of a powerful model, students experience the power of statistical techniques and learn why scientists require large numbers of individuals to zone in on a possible genomic location.
Later, when a disease breaks out on the island, students must put their knowledge about QTL analysis to work. The survival of dragons depends upon them! Students work to identify an area on a chromosome as the probable location of the disease gene and then hunt down the specific gene in a genetics database.
Computational genetics

In addition to teaching genetics concepts through a powerful model, GENIQUEST also introduces students to important tools and processes of computational genetics research. Researchers study mice as a “model organism” for humans because mice and humans are genetically 98% similar. Studying mice allows researchers to explore many models for human disease in cases where direct experimentation on humans is not possible. Likewise, “on the island,” experimentation on dragons is not permitted (in this fictional world, it takes 500 years for dragons to breed), so students must conduct research on another fictional animal, the drake.
To ensure that the underlying genome for both dragons and drakes was an accurate scientific analog, Jackson Lab researchers extended the original BioLogica genome by slicing the actual mouse genome into 50 parts, rearranging them randomly, and reassembling them with many genes removed. The resulting genome bears the same relationship to the mouse genome that our human genome does. Because it also has the same level of complexity as the mouse and human genomes—though with many fewer genes—computational biology techniques such as QTL analysis yield valid results in sizes that are manageable for students to explore.
With this enhanced BioLogica software, students learn the basics of QTL analysis and gain practice using the mouse database genome browser to research information about specific genes. When students pinpoint the probable disease location on a chromosome, they search for their answer within the actual Jackson Lab mouse gene database used by researchers worldwide.
Further study
With this background, students are prepared to embark on further study of real genetic data and scenarios. For instance, students might start with a QTL dataset that has already been published and test themselves, seeing if their analysis of the dataset leads them to the same genes that researchers identified as candidate genes for that dataset. Or they could analyze data from QTL analyses from numerous studies that have been publicly shared, but only partly analyzed.
Especially eager or advanced students could even run their own analysis on any of the huge number of public datasets that have not yet been tackled. This would offer the possibility of uncovering original research results, and require no more equipment than their standard computer. Magnet school students working with Jackson Lab regularly explore these possibilities, and advanced AP Biology students with a background from the GENIQUEST project would be well equipped to use it as well. With an understanding of biology concepts rooted in investigation of a robust biological model, students can approach biological problems on much firmer ground. Today’s GENIQUEST students might lead the way in tomorrow’s genetic research, unlocking mysteries important not only to dragons, but to humans as well.
What is QTL Analysis?
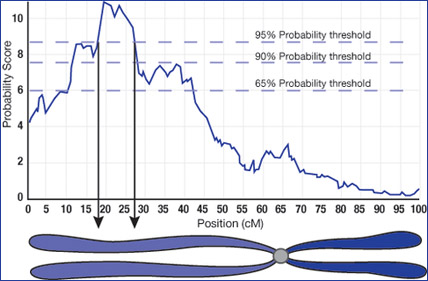
Genetics, history, and environment shape who we are. Specific traits like height, skin color, a tendency to obesity, or the development of diseases like diabetes can be a manifestation of genetic traits. The majority of these are “complex traits,” representing the interaction of many genes. Science and medical researchers are interested in determining what regions of the genome are linked to specific traits, especially traits for human diseases or conditions, for example, high cholesterol levels. One technique begins with organisms about which we already know much. Strains of mice are carefully bred so that every mouse of a certain strain is genetically identical. Specific locations on the genome are used to identify each strain. If we have one strain of mice that always has high cholesterol levels and another that always has low cholesterol levels, we can breed them and study the offspring. Because genetic crossover during breeding mixes the genes of the two strains, the offspring have mixed genomes. After breeding many offspring in a controlled way, the genomes of the resulting offspring represent a diverse mix of the two strains jumbled across all the specially identified locations on the genome. The next step is to study the offspring and their cholesterol levels and examine their “jumbled genomes” at each of the previously identified locations. If any of these locations are consistently associated with the original strain that always had high cholesterol and occur in individuals that have high cholesterol, it is very likely that genes in those locations have some control over the cholesterol levels. We use a statistical process known as Quantitative Trait Loci (QTL) analysis on the large population to make that association and, thus, to identify probable regions of the genome for further investigation.
SAIL
SAIL (Scalable Architecture for Interactive Learning) is both a framework and a collection of applications and databases that permit non-programmers to create, modify, and deploy from the Web dynamically created learning activities with embedded highly interactive simulation, modeling, probeware, graphing, and analysis components. All the work a learner does while interacting with a SAIL client-based activity—visiting pages, answering questions, creating a drawing, investigating a model, collecting data from probes—is saved over the network and is available for the learner when the activity is run again. In addition, the learner’s work is available for a teacher or researcher to review. The current SAIL framework, which is an outgrowth of the NSF-funded Web-based Inquiry Science Environment, has been developed by the NSF Center for Technology Enhanced Learning in Science (TELS) to support the development of inquiry learning investigations. SAIL represents over a decade of prior research into technology-enhanced student learning.